Detection of LytA Genes in Streptococcus pneumoniae Isolated from sputum pneumonia patients
Abstract
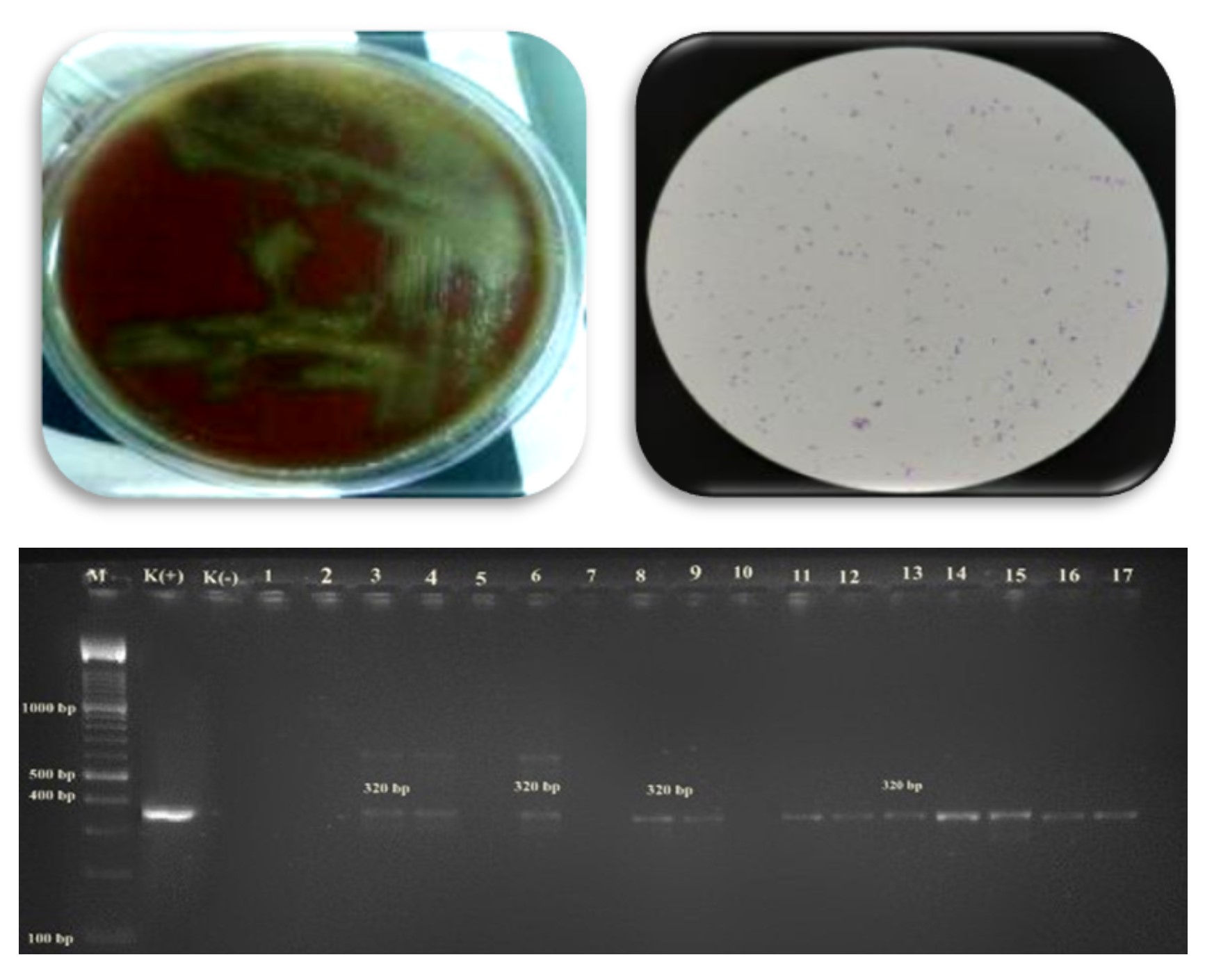
Streptococcus pneumoniae (pneumococcus) is a Gram-positive facultative anaerobic bacterium that is a major cause of morbidity and mortality worldwide. But the lack of reporting of disease by this bacterium in Indonesia, one of the causes is because the diagnosis of pneumococcal infection is often clinically not typical and conventional methods which are still the standard gold method often give false-negative results. So the purpose of this study was to evaluate the performance of culture and molecular diagnostic methods using the Polymerase Chain Reaction (PCR) technique in detecting Streptococcus pneumoniae in sputum clinical samples using the Autolysin (LytA) gene which is a virulence factor of this bacterium. 57 isolates from 60 samples were confirmed as Streptococcus sp through microscopic identification, culture, and biochemical tests. Then the sensitivity test with an optochin test of 9 (9%) compared the results descriptively with the PCR technique using the Autolysin A (LytA) gene which was obtained more sensitive by 15 (25%).
References
Balsells, E. (2017) ‘Serotype distribution of Streptococcus pneumoniae causing invasive disease in children in the post-PCV era: A systematic review and meta-analysis’, PLoS ONE. doi: 10.1371/journal.pone.0177113.
Bandettini, R. and Melioli, G. (2012) ‘Laboratory diagnosis of Streptococcus pneumoniae infections: Past and future’, Journal of Preventive Medicine and Hygiene. doi: 10.15167/2421-4248/jpmh2012.53.2.321.
Bogaert, D., De Groot, R. and Hermans, P. W. M. (2004) ‘Streptococcus pneumoniae colonisation: The key to pneumococcal disease’, Lancet Infectious Diseases. doi: 10.1016/S1473-3099(04)00938-7.
Chen, Yu Shen et al. (2013) ‘Comparison of diagnostic tools with multiplex polymerase chain reaction for pediatric lower respiratory tract infection: A single center study’, Journal of Microbiology, Immunology and Infection, 46(6), pp. 413–418. doi: 10.1016/j.jmii.2012.07.016.
Fischetti, V. A. and Ryan, P. (2015) ‘Streptococcus’, in Practical Handbook of Microbiology, Third Edition, pp. 99–114. doi: 10.1201/b17871.
Gholamhosseini-Moghaddam, T. et al. (2015) ‘Detection of LytA, pspC, and rrgA genes in Streptococcus pneumoniae isolated from healthy children’, Iranian Journal of Microbiology, 7(3), pp. 156–160.
Haas, W. et al. (2011) ‘High Proportion of Nontypeable Streptococcus pneumoniae Isolates among Sporadic, Nonoutbreak Cases of Bacterial Conjunctivitis’, Current Eye Research, 36(12), pp. 1078–1085. doi: 10.3109/02713683.2011.624670.
Lung, M. and Rello, J. (2014) ‘Microbiology of bacterial CAP using traditional and molecular techniques’, European Respiratory Monograph. doi: 10.1183/1025448x.10003213.
Maestro, B. and Sanz, J. M. (2016) ‘Choline binding proteins from Streptococcus pneumoniae: A dual role as enzybiotics and targets for the design of new antimicrobials’, Antibiotics. doi: 10.3390/antibiotics5020021.
Mousavi, S. F. et al. (2013) ‘Serotyping of Streptococcus pneumoniae isolated from Tehran by Multiplex PCR: Are serotypes of clinical and carrier isolates identical?’, Iranian Journal of Microbiology.
Prendki, V. et al. (2019) ‘Accuracy of comprehensive PCR analysis of nasopharyngeal and oropharyngeal swabs for CT-scan-confirmed pneumonia in elderly patients: a prospective cohort study’, Clinical Microbiology and Infection. doi: 10.1016/j.cmi.2018.12.037.Sampson, J. S. et al. (1997) ‘Limited diversity of Streptococcus pneumoniae psaA among pneumococcal vaccine serotypes’, Infection and Immunity. doi: 10.1128/iai.65.5.1967-1971.1997.
Scholz, C. F. P., Poulsen, K. and Kilian, M. (2012) ‘Novel molecular method for identification of Streptococcus pneumoniae applicable to clinical microbiology and 16S rRNA sequence-based microbiome studies’, Journal of Clinical Microbiology. doi: 10.1128/JCM.00365-12.
Simões, A. S. et al. (2016) ‘LytA-based identification methods can misidentify Streptococcus pneumoniae’, Diagnostic Microbiology and Infectious Disease, 85(2), pp. 141–148. doi: 10.1016/j.diagmicrobio.2016.03.018.
Varghese, R., Jayaraman, R. and Veeraraghavan, B. (2017) ‘Current challenges in the accurate identification of Streptococcus pneumoniae and its serogroups/serotypes in the vaccine era’, Journal of Microbiological Methods. doi: 10.1016/j.mimet.2017.07.015.
Wu, C. C. C. L. et al. (2015) ‘Recommendations for standards of monitoring during anaesthetsia and recovery (4th Ediction)’, Anesthesia and Analgesia. doi: 10.1186/1756-0500-6-276.
Zweifel, U. L. et al. (2016) ‘Preparation of McFarland Barium Sulfate Turbidity Standard (AMD-TP-02) in National Microbiology Standards’, Water Research. doi: 10.1373/clinchem.2008.112797.