Metagenomic Diversity of Gut Microbiota of Gestational Diabetes Mellitus of Pregnant Women
Abstract
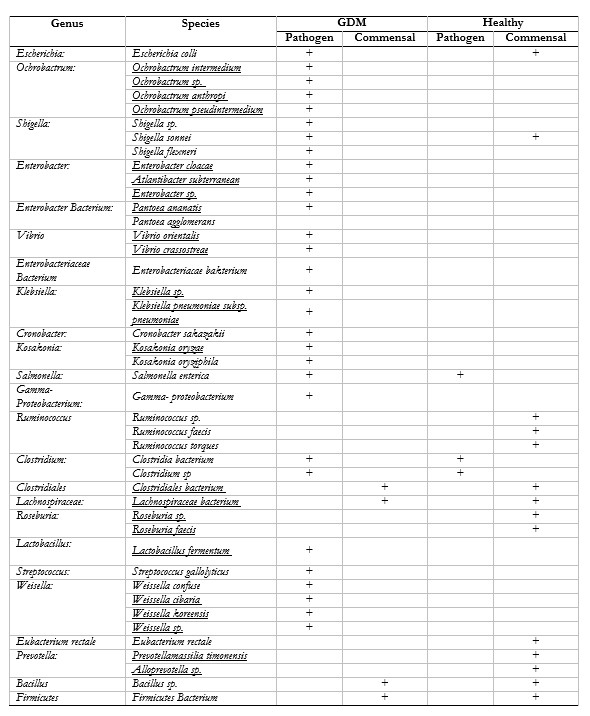
Gestational Diabetes Mellitus (GDM) is defined as a condition in which a woman without diabetes develops abnormal glucose tolerance that is first recognized during pregnancy. GDM is a significant public health problem with an incidence of 1.9 – 3.6% of all pregnancies in Indonesia. Additionally, women with GDM during pregnancy have a high risk of developing diabetes when they are not pregnant, such as type 2 diabetes (T2D). One alternative variable in the management of T2D globally is gut microbiota. Here, to find out the role of gut microbiota in pregnancy, we characterized the stools of 30 pregnant women, each consisting of fifteen GDM-detected pregnant women, and healthy pregnant women using metagenomic approach with genome analysis by directly isolating genomic DNA from the microbiota ecosystem that occupies the digestive tract. DNA sequencing results were analyzed by MEGA 6 software with the BLASTn algorithm in NCBI. Thus fifteen GDM-detected showed high nucleotide sequence homology with the Proteobacteria at phylum level, and Escherichia, Orchobacterium, Cronobacter, Shigella, Salmonella, Enterobacter, Klebsiella, Kosakonia, Vibrio dan Gamma-Proteobacterium at genus level compared to the healthy pregnant women which found by Firmicutes at phylum level and Ruminococcus, Clostridium, Clostridiales, Lachnospiraceae, Roseburia, Weisella, Eubacterium at genus level had a higher abundance in healthy pregnant women. In this result, we found also one of the fifteen healthy pregnant women showed differential abundance with enrichment of Prevotella species. Gut microbiota of GDM-diagnosed pregnant women has more varied composition, and dominated by the phylum Proteobacteria than in normal pregnant women.
References
Boerner, B. P. and Sarvetnick, N. E. (2011). Type 1 diabetes: Role of intestinal microbiome in humans and mice. Annals of the New York Academy of Sciences, 1243(1), pp. 103–118. doi: https://doi.org/10.1111/j.1749-6632.2011.06340.x.
Buchanan, T. A. Xiang, A., Kjos, S.L., and Watanabe, R. (2007). What is gestational diabetes? Diabetes Care, 30(SUPPL. 2). doi: https://doi.org/10.2337/dc07-s201.
Crusell, M. K. W., Hansen, T.H., Nielsen, T., Malte, C., Damm, P., Vestergaard, H., Rørbye, C., Jørgensen, N.R., Christiansen, O.B., Heinsen, F., Franke, A., Hansen, T., Lauenborg, J., and Pedersen, O. (2018). Gestational diabetes is associated with change in the gut microbiota composition in third trimester of pregnancy and postpartum. Microbiome. Microbiome, 6(1), p. 89. doi: https://doi.org/10.1186/s40168-018-0472-x.
Gomez-Arango, L. F. Barret, H.L., Mc-Lntyre, H.D., Callaway, L.K., Morrison M., and Nitert M.D. (2016). Connections between the gut microbiome and metabolic hormones in early pregnancy in overweight and obese women. Diabetes, 65(8), pp. 2214–2223. doi: https://doi.org/10.2337/db16-0278.
Ferrocino, I., Ponzo, V., Gambino, R., Zarovska, A., Leone, F., Monzeglio, C., Goitre, I., Rosato, R., Romano, A., Grassi, G., Broglio, F., Cassader, M., Cocolin, L., and Bo, S. (2018). Changes in the gut microbiota composition during pregnancy in patients with gestational diabetes mellitus (GDM). Scientific Reports. Springer US, 8(1), pp. 1–13. doi: https://doi.org/10.1038/s41598-018-30735-9.
Kamaruddin, M., Tokoro, M., Rahman, M.M., Arayama, S., Hidayati, A.P.N., Syafruddin, D., Asih, P.B.S., Yoshikawa, H., and Kawahara, E. (2014). Molecular characterization of various trichomonad species isolated from humans and related mammals in Indonesia’, Korean Journal of Parasitology, 52(5), pp. 471–478. doi: https://doi.org/10.3347/kjp.2014.52.5.471.
Koren, O., Goodrich, J.K., Cullender, T.C., Spor, A., Laitinen, K., Backhed, H.K., Gonzalez, A., Werner, J.J., Angenent, L.T., Knight, R., Backhed, F., Isolauri E., Salminen, S., and Ley, R.E. (2012). Host remodeling of the gut microbiome and metabolic changes during pregnancy. Cell, 150(3), pp. 470–480. doi: https://doi.org/10.1016/j.cell.2012.07.008.
Kamaruddin, M., Triananinsi, N., Sampara, N., Sumarni, Minarti, Rupa, A. M. (2020). Media kesehatan masyarakat. Media Kesehatan Masyarakat, 16(1), pp. 116–126. doi: http://dx.doi.org/10.30597/mkmi.v16i1.9050.
Qin, J., Raes, L.R., Arumugam M., Burgdorf, K.S., and Manichanh, C. (2010). A human gut microbial gene catalogue established by metagenomic sequencing. Nature. Nature Publishing Group, 464(7285), pp. 59–65. doi: https://doi.org/10.1038/nature08821.
Salzman, N. H., Hung, K., Haribhai, D., Chu, H., Karlsson-Sjoberg, J., Amir, E., Teggatz, P., Barman, M., Hayward, M., Eastwood, D., Stoel, M., Zhou, Y., Sodergen, E., Weinstock, G.M., Bevins, C.L., Williams, C.B., and Bos, N.A. (2010). Enteric defensins are essential regulators of intestinal microbial ecology HHS Public Access Author manuscript. Nat Immunol, 11(1), pp. 76–83. doi: https://doi.org/10.1038/ni.1825.
Saminan. (2012). Pertukaran Udara O2 dan CO2 dalam Pernapasan, Jurnal Kedokteran Syiah Kuala, 16, pp. 122–126. http://jurnal.unsyiah.ac.id/JKS/article/view/3509
Santacruz, A., Collado, M.C., Garcia-Valdes, L., Segura, M.T., Martin-Lagos, J.A., Anjos, T., Marti-Romero, M., Lopez, R.M., Florido, J., Campoy, C., and Sanz, Y. (2010). Gut microbiota composition is associated with body weight, weight gain and biochemical parameters in pregnant women. British Journal of Nutrition, 104(1), pp. 83–92. doi: https://doi.org/10.1017/S0007114510000176.
Soderborg, T. K., Borengasser, J., Barbour, L.A., and Friedman, J.E. (2016). Microbial transmission from mothers with obesity or diabetes to infants: an innovative opportunity to interrupt a vicious cycle. Diabetologia. Springer Verlag, pp. 895–906. doi: https://doi.org/10.1007/s00125-016-3880-0.
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution, 28(10), pp. 2731–2739. doi: https://doi.org/10.1093/molbev/msr121.
Wang, J., Zheng, J., Shi, W., Du, N., Xu, X., Zhang, Y., Ji, P., Zhang, F., Jia, Z., Wang, Y., Zheng, Z., Zhang, H., and Zhao, F. (2018). Dysbiosis of maternal and neonatal microbiota associated with gestational diabetes mellitus. Gut, 67(9), pp. 1614–1625. doi: https://doi.org/10.1136/gutjnl-2018-315988.
Wu, G. D., Chen, J., Hoffmann, C., Bittinger, K., Chen, Y-Y., Keilbaugh, S.A., Bewtra, M., Knights, D., Walters, W.A., Knight, R., Sinha, R., Gilroy, E., Gupta, K., Baldassano, R., Nessel, L., Li, H., Bushman, F.D., and Lewis, J.D.. (2011). Linking long-term dietary patterns with gut microbial enterotypes. Science, 334(6052), pp. 105–108. doi: https://doi.org/10.1126/science.1208344.